Last Updated on 01/12/2025 by Eran Feit
This tutorial provides a step-by-step guide on how to implement and train a U-Net model for polyp segmentation using TensorFlow/Keras.
The tutorial is divided into four parts:
🔹 Data Preprocessing and Preparation In this part, you load and preprocess the polyp dataset, including resizing images and masks, converting masks to binary format, and splitting the data into training, validation, and testing sets.
🔹 U-Net Model Architecture This part defines the U-Net model architecture using Keras. It includes building blocks for convolutional layers, constructing the encoder and decoder parts of the U-Net, and defining the final output layer.
🔹 Model Training Here, you load the preprocessed data and train the U-Net model. You compile the model, define training parameters like learning rate and batch size, and use callbacks for model checkpointing, learning rate reduction, and early stopping. The training history is also visualized.
🔹 Evaluation and Inference The final part demonstrates how to load the trained model, perform inference on test data, and visualize the predicted segmentation masks.
Check out our tutorial here : https://www.youtube.com/watch?v=YmWHTuefiws
Link for the full code here : https://eranfeit.lemonsqueezy.com/buy/34305755-4b6f-4db1-948b-e6d18c2aa640 or here : https://ko-fi.com/s/62988f9cc6
Link for my blog : https://eranfeit.net/blog/
You can find more tutorials, and join my newsletter here : https://eranfeit.net/
This tutorial is based on the U-net Architecture :
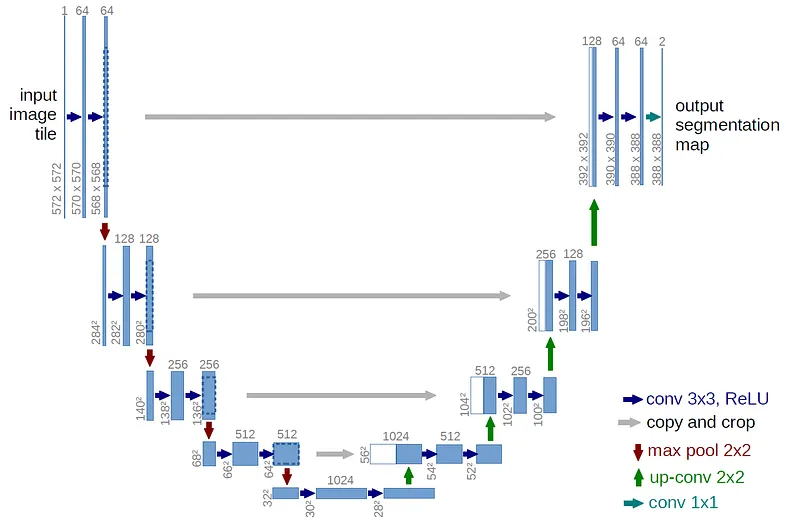
Here is the code for Polyp segmentation :
Link for the dataset : https://www.kaggle.com/datasets/balraj98/cvcclinicdb
Part 1: Image Preprocessing and Dataset Preparation for U-Net Model
This Python code preprocesses a dataset of polyp images and their corresponding segmentation masks.
It resizes the images, normalizes pixel values, converts segmentation masks to binary format, and splits the dataset into training, validation, and test sets.
Finally, it saves the preprocessed data as NumPy arrays for efficient loading and use in training a U-Net model for medical image segmentation.
# Dataset : https://www.kaggle.com/datasets/balraj98/cvcclinicdb
import cv2
import numpy as np
import os
# Define the dimensions to resize images and masks
height = 256
width = 256
# Initialize lists to store preprocessed images and masks
allImages = []
maskImages = []
# Paths to the dataset
path = "E:/Data-sets/Polyp/PNG/" # Base directory for the dataset
imagesPath = path + "Original" # Subdirectory for original images
maskPath = path + "Ground Truth" # Subdirectory for ground truth masks
# Count and print the number of images in the folder
print("Images in folder:")
images = os.listdir(imagesPath) # List all files in the image directory
print(len(images)) # Print the number of images
# Load a single image and its corresponding mask for inspection
img = cv2.imread(imagesPath + "/1.png", cv2.IMREAD_COLOR) # Load image in color mode
img = cv2.resize(img, (width, height)) # Resize image to 256x256
mask = cv2.imread(maskPath + "/1.png", cv2.IMREAD_GRAYSCALE) # Load mask in grayscale mode
mask = cv2.resize(mask, (width, height)) # Resize mask to 256x256
# Display the loaded image and mask
cv2.imshow("img", img)
cv2.imshow("mask", mask)
# Analyze the values in the resized mask for understanding
resizeto16 = cv2.resize(mask, (16, 16)) # Temporarily resize the mask to 16x16 for analysis
print(resizeto16) # Print the small-sized mask values
# Convert mask values to binary (0 for background, 1 for regions of interest)
resizeto16[resizeto16 <= 50] = 0 # Set pixel values <= 50 to 0
resizeto16[resizeto16 > 50] = 1 # Set pixel values > 50 to 1
print(resizeto16) # Print the binary mask
cv2.waitKey(0) # Wait for a key press before closing the displayed images
# Preprocess all images and masks in the dataset
for imagefile in images:
# Preprocess the image
file = imagesPath + "/" + imagefile # Construct the full file path
img = cv2.imread(file, cv2.IMREAD_COLOR) # Load the image in color mode
img = cv2.resize(img, (width, height)) # Resize the image to 256x256
img = img / 255.0 # Normalize pixel values to the range [0, 1]
img = img.astype(np.float32) # Convert the image to float32 type
allImages.append(img) # Add the processed image to the list
# Preprocess the mask
file = maskPath + "/" + imagefile # Construct the full mask file path
mask = cv2.imread(file, cv2.IMREAD_GRAYSCALE) # Load the mask in grayscale mode
mask = cv2.resize(mask, (width, height)) # Resize the mask to 256x256
mask[mask <= 50] = 0 # Set pixel values <= 50 to 0 (background)
mask[mask > 50] = 1 # Set pixel values > 50 to 1 (foreground)
maskImages.append(mask) # Add the processed mask to the list
# Convert lists to NumPy arrays for efficient computation
allImagesNP = np.array(allImages) # Convert the images list to a NumPy array
maskImageNP = np.array(maskImages) # Convert the masks list to a NumPy array
maskImageNP = maskImageNP.astype(int) # Ensure masks have integer values (0 or 1)
# Print the shapes and data types of the arrays for verification
print(allImagesNP.shape) # Shape of the images array
print(allImagesNP.dtype) # Data type of the images array
print(maskImageNP.shape) # Shape of the masks array
print(maskImageNP.dtype) # Data type of the masks array
# Split the dataset into training, validation, and test sets
from sklearn.model_selection import train_test_split
# Split 90% for training and 10% for testing
X_train, X_test, y_train, y_test = train_test_split(allImagesNP, maskImageNP, test_size=0.1, random_state=42)
# Further split the training set into 80% training and 10% validation
X_train, X_val, y_train, y_val = train_test_split(X_train, y_train, test_size=0.1, random_state=42)
# Print the shapes of the training, validation, and test datasets
print("X_train , X_val , y_train , x_val, X_test , y_test ------> shapes :")
print(X_train.shape)
print(y_train.shape)
print(X_val.shape)
print(y_val.shape)
print(X_test.shape)
print(y_test.shape)
# Save the processed datasets as NumPy files for reuse
print("Start saving the data:")
np.save("e:/temp/Unet-Polip-images-X_train.npy", X_train) # Save training images
np.save("e:/temp/Unet-Polip-images-y_train.npy", y_train) # Save training masks
np.save("e:/temp/Unet-Polip-images-X_val.npy", X_val) # Save validation images
np.save("e:/temp/Unet-Polip-images-y_val.npy", y_val) # Save validation masks
np.save("e:/temp/Unet-Polip-images-X_test.npy", X_test) # Save test images
np.save("e:/temp/Unet-Polip-images-y_test.npy", y_test) # Save test masks
print("Finished saving the data.")Link for the full code here : https://ko-fi.com/s/62988f9cc6
Part 2 : Create the U-Net building blocks
(U-Net Architecture Implementation for Image Segmentation )
This Python code defines a U-Net model for image segmentation tasks using TensorFlow and Keras. It includes convolutional blocks, an encoder-decoder structure with skip connections, and a sigmoid-activated output layer suitable for binary segmentation. The model architecture is modular and parameterized to adapt to different input shapes.
import tensorflow as tf
from tensorflow.keras.layers import * # Importing essential layers from Keras
from tensorflow.keras.models import Model # Importing the functional API for model creation
# Define a convolutional block with two convolutional layers, batch normalization, and ReLU activation
def conv_block(x, num_filters):
# First convolutional layer
x = Conv2D(num_filters, (3, 3), padding="same")(x) # Apply a 3x3 convolution
x = BatchNormalization()(x) # Normalize the batch to stabilize training
x = Activation("relu")(x) # Apply ReLU activation
# Second convolutional layer
x = Conv2D(num_filters, (3, 3), padding="same")(x) # Another 3x3 convolution
x = BatchNormalization()(x) # Batch normalization again
x = Activation("relu")(x) # ReLU activation
return x # Return the processed tensor
# Define the U-Net model
def build_model(shape):
"""
Build a U-Net model for binary image segmentation.
Parameters:
- shape: Tuple representing the input image dimensions, e.g., (256, 256, 3).
Returns:
- A TensorFlow Keras model.
"""
# Define the number of filters for each level of the U-Net
num_filters = [16, 32, 48, 64] # Increasing filters for deeper levels
# Input layer
inputs = Input((shape)) # Define the input tensor based on the given shape
# Initialize a list to store skip connections
skip_x = []
x = inputs # Start with the input tensor
# Encoder part of the U-Net
for f in num_filters:
x = conv_block(x, f) # Apply the convolutional block
skip_x.append(x) # Store the output for skip connections
x = MaxPool2D((2, 2))(x) # Downsample the feature map using max pooling
# Bridge between encoder and decoder with 128 filters
x = conv_block(x, 128) # Convolutional block at the bottleneck
# Prepare for the decoder by reversing the filter sizes and skip connections
num_filters.reverse() # Reverse the order of filters for decoding
skip_x.reverse() # Reverse the skip connections for decoding
# Decoder part of the U-Net
for i, f in enumerate(num_filters):
x = UpSampling2D((2, 2))(x) # Upsample the feature map to match the skip connection
xs = skip_x[i] # Retrieve the corresponding skip connection
x = Concatenate()([x, xs]) # Concatenate the upsampled tensor with the skip connection
x = conv_block(x, f) # Apply a convolutional block
# Output layer
x = Conv2D(1, (1, 1), padding="same")(x) # Apply a 1x1 convolution to produce a single-channel output
x = Activation("sigmoid")(x) # Apply sigmoid activation for binary classification (segmentation)
return Model(inputs, x) # Return the complete U-Net modelLink for the full code here : https://ko-fi.com/s/62988f9cc6
Part 3 : Model Training and Evaluating a U-Net Model for Polyp Segmentation
This Python script trains a U-Net model for binary image segmentation of polyps using preprocessed data.
It uses TensorFlow and Keras to build the model, compiles it with an Adam optimizer and binary cross-entropy loss, and employs callbacks for model checkpointing, learning rate reduction, and early stopping.
The training progress is visualized with accuracy and loss plots, and the model’s performance is evaluated on a test set.
import numpy as np
# Load preprocessed datasets for training, validation, and testing
X_train = np.load("e:/temp/Unet-Polip-images-X_train.npy") # Training images
y_train = np.load("e:/temp/Unet-Polip-images-y_train.npy") # Training masks
X_val = np.load("e:/temp/Unet-Polip-images-X_val.npy") # Validation images
y_val = np.load("e:/temp/Unet-Polip-images-y_val.npy") # Validation masks
X_test = np.load("e:/temp/Unet-Polip-images-X_test.npy") # Test images
y_test = np.load("e:/temp/Unet-Polip-images-y_test.npy") # Test masks
# Print the shapes of the loaded datasets for verification
print(X_train.shape) # Shape of training data
print(y_train.shape) # Shape of training masks
print(X_val.shape) # Shape of validation data
print(y_val.shape) # Shape of validation masks
print(X_test.shape) # Shape of test data
print(y_test.shape) # Shape of test masks
# Define input image dimensions
Height = 256
Width = 256
# Import TensorFlow and necessary functions for building and training the model
import tensorflow as tf
from Step02BuildTheUnetModel import build_model # Custom function to build the U-Net model
from keras.callbacks import ModelCheckpoint, ReduceLROnPlateau, EarlyStopping
# Define model parameters
shape = (256, 256, 3) # Input image shape
lr = 1e-4 # Learning rate for the optimizer
batch_size = 8 # Batch size for training
epochs = 50 # Number of training epochs
# Build the U-Net model
model = build_model(shape) # Initialize the U-Net model
#print(model.summary()) # Uncomment to display the model's architecture
# Compile the model with the Adam optimizer and binary cross-entropy loss
opt = tf.keras.optimizers.Adam(lr)
model.compile(loss="binary_crossentropy", optimizer=opt, metrics=['accuracy'])
# Calculate the number of steps per epoch for training and validation
stepsPerEpoch = np.ceil(len(X_train) / batch_size)
validationSteps = np.ceil(len(X_val) / batch_size)
# Define the file path to save the best model
best_model_file = "e:/temp/PolypSegment.h5"
# Define callbacks for training
callbacks = [
ModelCheckpoint(best_model_file, verbose=1, save_best_only=True), # Save the best model
ReduceLROnPlateau(monitor="val_loss", patience=3, factor=0.1, verbose=1, min_lr=1e-6), # Reduce learning rate on plateau
EarlyStopping(monitor="val_loss", patience=5, verbose=1) # Stop training early if no improvement
]
# Train the model with the training and validation datasets
history = model.fit(
X_train, y_train,
batch_size=batch_size,
epochs=epochs,
verbose=1,
validation_data=(X_val, y_val),
validation_steps=validationSteps,
steps_per_epoch=stepsPerEpoch,
shuffle=True,
callbacks=callbacks
)
# Import Matplotlib for visualizing training progress
import matplotlib.pyplot as plt
# Retrieve training history for accuracy and loss
acc = history.history['accuracy'] # Training accuracy
val_acc = history.history['val_accuracy'] # Validation accuracy
loss = history.history['loss'] # Training loss
val_loss = history.history['val_loss'] # Validation loss
# Get the range of epochs
epochs = range(len(acc))
# Plot training and validation accuracy
plt.plot(epochs, acc, 'r', label="Train accuracy")
plt.plot(epochs, val_acc, 'b', label="Validation accuracy")
plt.xlabel('Epoch')
plt.ylabel('Accuracy')
plt.title("Train and Validation Accuracy")
plt.legend(loc='lower right')
plt.show()
# Plot training and validation loss
plt.plot(epochs, loss, 'r', label="Train loss")
plt.plot(epochs, val_loss, 'b', label="Validation loss")
plt.xlabel('Epoch')
plt.ylabel('Loss')
plt.title("Train and Validation Loss")
plt.legend(loc='upper right')
plt.show()
# Evaluate the model on the test dataset
resultEval = model.evaluate(X_test, y_test)
print("Evaluate the test data:")
print(resultEval) # Print the evaluation metrics (loss and accuracy)Link for the full code here : https://ko-fi.com/s/62988f9cc6
Part 4 : Loading and Testing a U-Net Model for Polyp Segmentation
This Python script demonstrates how to load a trained U-Net model, evaluate it on test data, and visualize the segmentation result for a selected test image.
The model predicts a binary segmentation mask, with pixel values thresholded to distinguish between the background (black) and the segmented region (white).
The visualization uses OpenCV to display the original test image and the predicted mask.
import numpy as np
import tensorflow as tf
import cv2
# Path to the saved trained model
best_model_file = "e:/temp/PolypSegment.h5"
# Load the trained U-Net model
model = tf.keras.models.load_model(best_model_file)
#print(model.summary()) # Uncomment to display the model's architecture
# Load the test dataset
X_test = np.load("e:/temp/Unet-Polip-images-X_test.npy") # Test images
y_test = np.load("e:/temp/Unet-Polip-images-y_test.npy") # Test masks
# Select a specific test image to visualize
img = X_test[4] # Selecting the 5th test image (index 4)
# Display the original image (optional)
#cv2.imshow("img", img) # Uncomment to view the image
#cv2.waitKey(0) # Wait for a key press to close the window
# Prepare the test image for model prediction
imgForModel = np.expand_dims(img, axis=0) # Add a batch dimension to match model input shape
# Print the shapes for verification
print(img.shape) # Shape of the selected test image
print(imgForModel.shape) # Shape of the image after adding batch dimension
# Perform prediction using the trained model
p = model.predict(imgForModel) # Predict the segmentation mask
result = p[0] # Extract the predicted mask from the batch
# Print the shape of the predicted result
print(result.shape)
# Post-process the predicted mask
# For binary classification: values above 0.5 are set to 255 (white), others to 0 (black)
result[result <= 0.5] = 0 # Background
result[result > 0.5] = 255 # Segmented region
# Display the original image and the predicted mask
cv2.imshow("Original Image", img) # Show the test image
cv2.imshow("Predicted Mask", result) # Show the predicted binary mask
cv2.waitKey(0) # Wait for a key press to close the windows
cv2.destroyAllWindows() # Close all OpenCV windowsLink for the full code here : https://ko-fi.com/s/62988f9cc6
Here is result :
This is our test image and the mask for the polyp segmentation :

Connect :
☕ Buy me a coffee — https://ko-fi.com/eranfeit
🖥️ Email : feitgemel@gmail.com
🤝 Fiverr : https://www.fiverr.com/s/mB3Pbb
Planning a trip and want ideas you can copy fast?
Here are three detailed guides from our travels:
• 5-Day Ireland Itinerary: Cliffs, Castles, Pubs & Wild Atlantic Views
https://eranfeit.net/unforgettable-trip-to-ireland-full-itinerary/
• My Kraków Travel Guide: Best Places to Eat, Stay & Explore
https://eranfeit.net/my-krakow-travel-guide-best-places-to-eat-stay-explore/
• Northern Greece: Athens, Meteora, Tzoumerka, Ioannina & Nafpaktos (7 Days)
https://eranfeit.net/my-amazing-trip-to-greece/
Each guide includes maps, practical tips, and family-friendly stops—so you can plan in minutes, not hours.
Enjoy,
Eran
